Prior Work#
Prior to our work, research on deep-learning-based EEG decoding was limited
Few studies compared to published feature-based decoding results
Most EEG DL architectures had only 1-3 convolutional layers and included fully-connected layers with many parameters
Most work only considered very restricted frequency ranges
Most studies only compared few design choices and training strategies
Prior to 2017, when the first work presented in this thesis was published, there was only limited literature on EEG decoding with deep learning. In this chapter, I outline what decoding problems, input representations, network architectures, hyperparameter choices and visualizations were evaluated in prior work. This is based on the literature research that we presented in Schirrmeister et al. [2017].
Decoding Problems and Baselines#
Decoding problem |
Number of studies |
With published baseline |
|---|---|---|
Imagined or Executed Movement |
6 |
2 |
Oddball/P300 |
5 |
1 |
Epilepsy-related |
4 |
2 |
Music Rhythm |
2 |
0 |
Memory Performance/Cognitive Load |
2 |
0 |
Driver Performance |
1 |
0 |
The most widely studied decoding problems were movement-related decoding problems such as decoding which body part (hand, feet etc.) a person is moving or imagining to move (see Table 1). From the 19 studies we identified at the time, only 5 compared their decoding results to an external published baseline result, limiting the insights about deep-learning EEG decoding performance. We therefore decided to compare deep-learning EEG decoding to a strong feature-based baseline (see Filter Bank Common Spatial Patterns and Filterbank Network) on widely researched movement-related decoding tasks.
Input Domains and Frequency Ranges#
Show code cell source
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import cm
import seaborn
import numpy as np
import re
from myst_nb import glue
seaborn.set_palette('colorblind')
seaborn.set_style('darkgrid')
import re
%matplotlib inline
%config InlineBackend.figure_format = 'png'
#matplotlib.rcParams['figure.figsize'] = (12.0, 1.0)
matplotlib.rcParams['font.size'] = 14
a = np.array(['Time, 8–30 Hz ', 'Time, 0.1–40 Hz ', 'Time, 0.05–15 Hz ',
'Time, 0.3–20 Hz ', 'Frequency, 6–30 Hz ', ' Frequency, 0–200 Hz ',
'Time, 1–50 Hz ', ' Time, 0–100 HZ ',
'Frequency, mean amplitude for 0–7 Hz, 7–14 Hz, 14–49 Hz ',
'Time, 0.5–50 Hz ', 'Time, 0–128 Hz ',
' Frequency, mean power for 4–7 Hz, 8–13 Hz, 13–30 Hz ',
'Time, 0.5–30Hz ', 'Time, 0.1–50 Hz ',
'Frequency, 4–40 Hz, using FBCSP ',
' Time and frequency evaluated, 0-200 Hz ', 'Frequency, 8–30 Hz ',
'Time, 0.15–200 Hz ', ' Time, 0.1-20 Hz '])
domain_strings = [s.split(',')[0] for s in a]
start_fs = [float(re.sub(r'[a-z ]+',r'', re.split(r'[–-–-]'," ".join(s.split(',')[1:]))[0])) for s in a]
end_fs = [float(re.sub(r'[a-z HZFBCSP]+',r'', re.split(r'[–-–-]'," ".join(s.split(',')[1:]))[1])) for s in a]
domain_strings = np.array(domain_strings)
start_fs = np.array(start_fs)
end_fs = np.array(end_fs)
freq_mask = np.array(['freq' in s.lower() for s in domain_strings])
time_mask = np.array(['time' in s.lower() for s in domain_strings])
fig = plt.figure(figsize=(8,4))
rng = np.random.RandomState(98349384)
color = seaborn.color_palette()[0]
i_sort = np.flatnonzero(time_mask)[np.argsort(end_fs[time_mask])]
for i, (d,s,e) in enumerate(zip(
domain_strings[i_sort], start_fs[i_sort], end_fs[i_sort])):
offset = 0.6*i/len(i_sort) - 0.3
plt.plot([offset,offset] , [s, e], marker='o', alpha=1, color=color, ls='-')
i_sort = np.flatnonzero(freq_mask)[np.argsort(end_fs[freq_mask])]
for i, (d,s,e) in enumerate(zip(
domain_strings[i_sort], start_fs[i_sort], end_fs[i_sort])):
offset = 0.6*i/len(i_sort) + 0.7
plt.plot([offset,offset] , [s, e], marker='o', alpha=1, color=color, ls='-')
plt.xlim(-0.5,1.5)
plt.xlabel("Input domain")
plt.ylabel("Frequency [Hz]")
plt.xticks([0,1], ["Time", "Frequency"], rotation=45)
plt.title("Input domains and frequency ranges in prior work", y=1.05)
plt.yticks([0,25,50,75,100,150,200])
glue('input_domain_fig', fig)
plt.close(fig)
None
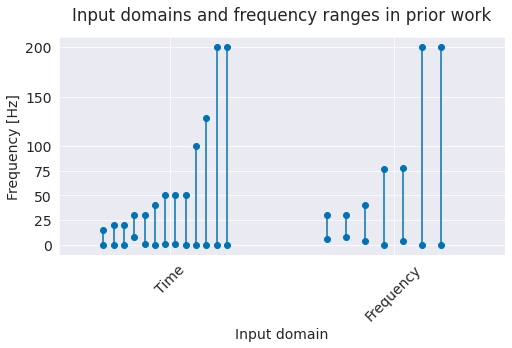
Fig. 1 Input domains and frequency ranges in prior work. Grey lines represent frequency ranges of individual studies. Note that many studies only include frequencies below 50 Hz, some use very restricted ranges (alpha/beta band).#
Deep networks can either decode directly from the time-domain EEG or process the data in the frequency domain, for example after a Fourier transformation. 12 of the prior studies used time-domain inputs, 6 used frequency-domain inputs and one used both. We decided to work directly in the time domain, as the deep networks should in principle be able to learn how to extract any needed spectral information from the time-domain input.
Most prior studies that were working in the time domain only used frequencies below 50 Hz. We were interested in how well deep networks can also extract lesser-used higher-frequency components of the EEG signal. For that, we used a sampling rate of 250 Hz, which means we were able to analyze frequencies up to the Nyquist frequency of 125 Hz. As a suitable dataset where high-frequency information may help decoding, we included our high-gamma dataset in our study, since it was recorded specifically to allow extraction of higher-frequency (>50 Hz) information from scalp EEG [Schirrmeister et al., 2017].
Network Architectures#
Show code cell source
ls = np.array([' 2/2 ', ' 3/1 ', ' 2/2 ', ' 3/2 ', ' 1/1 ', ' 1/2 ', ' 1/3 ',
' 1–2/2 ', ' 3/1 (+ LSTM as postprocessor) ', ' 4/3 ', ' 1-3/1-3 ',
' 3–7/2 (+ LSTM or other temporal post-processing (see design choices)) ',
' 2/1 ', ' 3/3 (Spatio-temporal regularization) ',
' 2/2 (Final fully connected layer uses concatenated output by convolutionaland fully connected layers) ',
' 1-2/1 ',
'2/0 (Convolutional deep belief network, separately trained RBF-SVM classifier) ',
' 3/1 (Convolutional layers trained as convolutional stacked autoencoder with target prior) ',
' 2/2 '])
conv_ls = [l.split('/')[0] for l in ls]
low_conv_ls = [int(re.split(r'[–-]', c)[0])for c in conv_ls]
high_conv_ls = [int(re.split(r'[–-]', c)[-1])for c in conv_ls]
dense_ls = [l.split('/')[1] for l in ls]
low_dense_ls = [int(re.split(r'[–-]', c[:8])[0][:2])for c in dense_ls]
high_dense_ls = [int(re.split(r'[–-]', c[:8])[-1][:2])for c in dense_ls]
all_conv_ls = np.concatenate([np.arange(low_c, high_c+1) for low_c, high_c in zip(low_conv_ls, high_conv_ls)])
all_dense_ls = np.concatenate([np.arange(low_c, high_c+1) for low_c, high_c in zip(low_dense_ls, high_dense_ls)])
bincount_conv = np.bincount(all_conv_ls)
bincount_dense = np.bincount(all_dense_ls)
rng = np.random.RandomState(98349384)
color = seaborn.color_palette()[0]
fig = plt.figure(figsize=(8,4))
for low_c, high_c in zip(low_conv_ls, high_conv_ls):
offset = rng.randn(1) * 0.1
tried_cs = np.arange(low_c, high_c+1)
plt.plot([offset,] * len(tried_cs), tried_cs, marker='o', alpha=0.5, color=color, ls=':')
for i_c, n_c in enumerate(bincount_conv):
plt.scatter(0.4, i_c, color=color, s=n_c*40)
plt.text(0.535, i_c, str(n_c)+ "x", ha='left', va='center')
for low_c, high_c in zip(low_dense_ls, high_dense_ls):
offset = 1 + rng.randn(1) * 0.1
tried_cs = np.arange(low_c, high_c+1)
plt.plot([offset,] * len(tried_cs), tried_cs, marker='o', alpha=0.5, color=color, ls=':')
for i_c, n_c in enumerate(bincount_dense):
plt.scatter(1.4, i_c, color=color, s=n_c*40)
plt.text(1.535, i_c, str(n_c)+ "x", ha='left', va='center')
plt.xlim(-0.5,2)
plt.xlabel("Type of layer")
plt.ylabel("Number of layers")
plt.xticks([0,1], ["Convolutional", "Dense"], rotation=45)
plt.yticks([1,2,3,4,5,6,7]);
plt.title("Number of layers in prior works' architectures", y=1.05)
glue('layernum_fig', fig)
plt.close(fig)
None
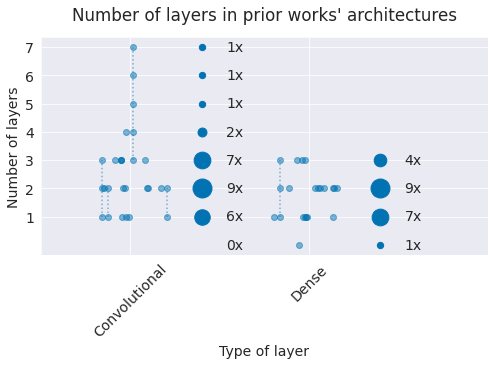
Fig. 2 Number of layers in prior work. Small grey markers represent individual architectures. Dashed lines indicate different number of layers investigated in a single study (e.g., a single study investigated 3-7 convolutional layers). Larger grey markers indicate sum of occurences of that layer number over all studies (e.g., 9 architectures used 2 convolutional layers). Note most architectures use only 1-3 convolutional layers.#
The architectures used in prior work typically only included up to 3 layers, with only 2 studies considering more layers. As network architectures in other domains tend to be a lot deeper, we also evaluated architectures with a larger number of layers in our work. Several architectures from prior work also included fully-connected layers with larger number of parameters which had fallen out of favor in computer-vision deep-learning architectures due to their large compute and memory requirements with little accuracy benefit. Our architectures do not include traditional fully-connected layers with a large number of parameters.
Hyperparameter Evaluations#
Study |
Design choices |
Training strategies |
|---|---|---|
Kernel sizes |
||
Different time windows |
||
Addition of six-layer stacked autoencoder on ConvNet features |
||
Different subdivisions of frequency range |
||
Replacement of convolutional layers by restricted Boltzmann machines with slightly varied network architecture} |
||
1 or 2 convolutional layers |
||
Cross-subject supervised training, within-subject finetuning of fully connected layers |
||
Number of convolutional layers |
||
Kernel sizes |
Pretraining first layer as convolutional autoencoder with different constraints |
|
Combination ConvNet and MLP (trained on different features) vs. only ConvNet vs. only MLP |
||
Best values from automatic hyperparameter optimization: frequency cutoff, one vs two layers, kernel sizes, number of channels, pooling width |
Best values from automatic hyperparameter optimization: learning rate, learning rate decay, momentum, final momentum |
|
Partially supervised CSA |
||
Electrode subset (fixed or automatically determined) |
Prior work varied widely in their comparison of design choices and training strategies. 6 of the studies did not compare any design choices or training strategy hyperparameters. The other 13 studies evaluated different hyperparameters, with the most common one the kernel size (see Table 2). Only one study evaluated a wider range of hyperparameters [Stober et al., 2014]. To fill this gap, we compared a wider range of design choices and training strategies and specifically evaluated whether improvements of computer vision architecture design choices and training strategies also lead to improvements in EEG decoding.
Visualizations#
Study |
Visualization type(s) |
Visualization findings |
|---|---|---|
Weights (spatial) |
Largest weights found over prefrontal and temporal cortex |
|
Weights |
Weights showed typical P300 distribution |
|
Weights (spatial + frequential) |
Some weights represented difference of values of two electrodes on different sides of head |
|
Weights |
Clusters of weights showed typical frequency band subdivision (delta, theta, alpha, beta, gamma) |
|
Weights |
Weights increasingly correlated with IED waveforms with increasing number of training iterations |
|
Input occlusion and effect on prediction accuracy |
Allowed to locate areas critical for seizure |
|
Weights (spatial) |
Some filter weights had expected topographic distributions for P300 |
|
Inputs that maximally activate given filter |
Different filters were sensitive to different frequency bands |
|
Weights (spatial+3 timesteps, pretrained as autoencoder) |
Different constraints led to different weights, one type of constraints could enforce weights that are similar across subjects; other type of constraints led to weights that have similar spatial topographies under different architectural configurations and preprocessings |
|
Weights |
Spatiotemporal regularization led to softer peaks in weights |
|
Weights |
Spatial filters were similar for different architectures |
Visualizations can help understand what information the networks are extracting from the EEG signal. 11 of the prior 19 studies presented any visualizations. These studies mostly focused on analyzing weights and activations, see Table 3. In our work, we first focused on investigating how far the networks extract spectral features known to work well for movement-related decoding, see Perturbation Visualization. Later, we also developed more sophisticated visualization methods and applied them both to pathology decoding, see Invertible Networks and Understanding Pathology Decoding With Invertible Networks.
Open Questions
How do ConvNets perform on well-researched EEG movement-related decoding tasks against strong feature-based baselines?
How do shallower and deeper architectures compare?
How do design choices and training strategies affect the decoding performance?
What features do the deep networks learn on the EEG signals?
Do they learn to use higher-frequency (>50 Hz) information?